Non-coding RNA & inflammation
The advent of deep sequencing methods in the past decade has revealed the pervasive transcription of mammalian genomes. As a result thousands of previously unknown non-coding RNA genes have been discovered. Most recently long non-coding RNAs (lncRNAs) have been described, which resemble mRNAs but lack protein-coding potential. LncRNAs are belived to regulate a variety of cellular processes by acting as scaffolds for protein complex assembly or decoys and guides controling the function of individual proteins.
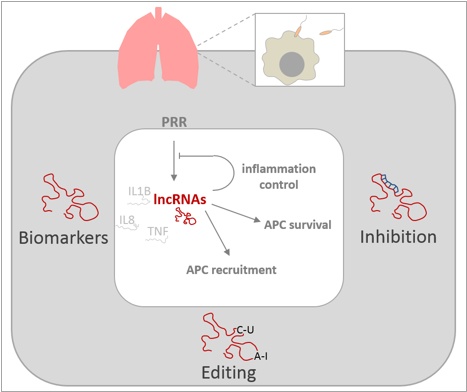
Our group studies the involvement of lncRNAs and other emerging non-coding RNA species in inflammatory responses to bacterial pathogens. Particularly we adress the roles of lncRNAs in human lung innate immune responses. We explore the involvement of lncRNAs in adequate activation of antigen presenting cells (APCs) of the lung and adress their roles in the interaction of APCs with the lung epithelium. To this end we employ a variety of methods ranging from RNA-Seq to genome editing and RNA affinity purification. Our ongoing projects on human lung APC and epithelial immunity are detailed below.

Antigen presenting cells
Alveolar macrophages constitute a population of tissue resident antigen-presenting cells with major roles in the orchestration of local inflammatory responses and tissue repair. We colaborate with clinicians to collect alveolar macrophages from patients and healthy volonteers and characterize the transcriptional profiles of these cells under homeostatic and inflammatory conditions. We are specifically interested in the roles of lncRNAs in alveolar macrophage immune responses to microbial pathogens and use both primary cells and cell-line models to reveal lncRNA loss-of-funtion phenotypes. Through colaborations we are furthermore adressing whether lncRNAs may constitute novel therapeutic targets for modulation of APC activity using antisense chemistry.
Strategies of bacterial and viral infectious agents
In addition to the role of RNA in the immune system, we investigate non-coding RNA mechanisms and virulence strategies of infectious agents. In particular, we use high-throughput sequencing methods such as dual RNA-seq or differential RNA-seq to map the expression of bacterial genes in the human host and the architecture of bacterial virulence-associated operons and regulons. In addition to the disease-causing strategies of Salmonella, we study the behaviour of Legionella, Klebsiella and Vibrio in the human host and dissect their virulence gene repertoirs within the LOEWE Diffusible Signals network. In addition, we conduct studies on infections with influenza A and SARS-CoV-2 viruses.
Tools
Besides our above delineated projects we colaborate with chemists, pharmacists and engineers to develop tools which will facilitate the discovery of human ncRNA functions. Among others we work on improved CRISPR-based methods for ncRNA loss-of-function studies, improved antisense chemistry for lncRNA inhibition and bioartificial 3D tissue constructs. The latter allows us to investigate the roles of lncRNAs in critical intercellular communication axis coordinating the resident APC, PBMC, epithelial and endothelial cell responses during human lung inflammation. We also use our human lung tissue models as a platform to test potential therapeutic lncRNA inhibition approaches involving novel chemical modifications of PNA and LNA based inhibitors to confer cell-permeability. As a readout we monitor the inflammatory status of our constructs on knockdown of individual lncRNA candidates with PNA and LNA drugs.
We inform about recent achievements on our news page …