We’re excited to share that our latest research has been published in Nature Communications!
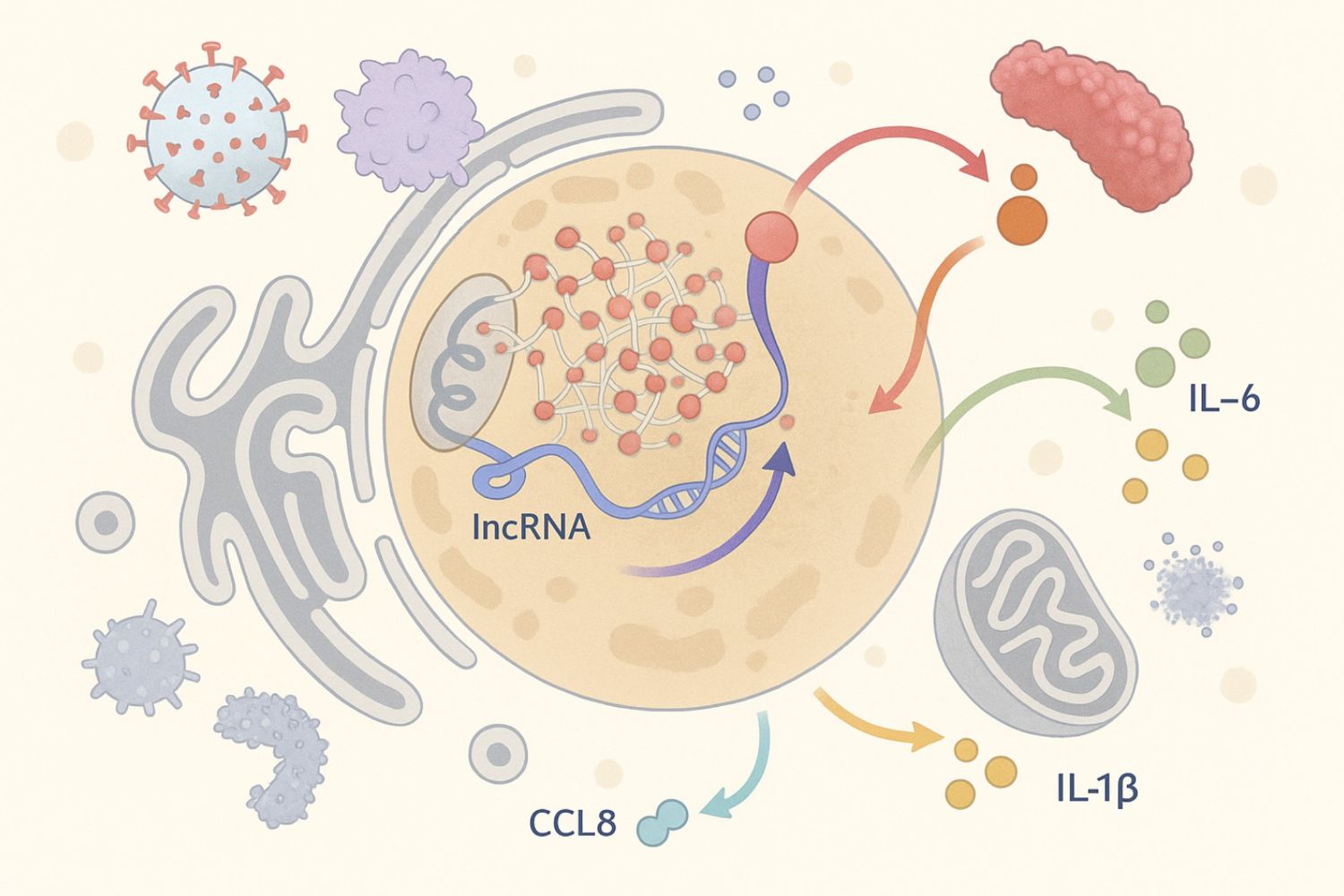
In this study, we set out to explore how long noncoding RNAs (lncRNAs) help immune cells—specifically human macrophages—sense and respond to bacterial infection.
To do this, we built a comprehensive multi-omics framework that integrates RNA-seq, proteomics, CRISPR interference screening, and RNA interactome profiling. The result is a transcriptome-wide atlas of immune-regulated lncRNAs, highlighting dozens of previously uncharacterized regulators of innate immunity.
A major highlight of our work is GRADR—a new method we developed to systematically predict protein partners of lncRNAs. GRADR combines gradient profiling with RNA-binding proteomics to reconstruct lncRNA–protein interactomes at scale. We used it to show that key immune-regulatory lncRNAs, including ROCKI, LINC01215, and AC022816.1, engage specific protein machineries such as splicing factors and nuclear scaffolds.
We also built SMyLR, the Searchable Myeloid lncRNA Registry (rna-lab.org/smylr), a free online tool that lets researchers explore the regulation, pathway dependency, and interactomes of hundreds of lncRNAs in human macrophages.
You can read the full paper here:
https://www.nature.com/articles/s41467-025-60084-x
Try out the SMyLR resource:
https://rna-lab.org/smylr

Comments are closed